- Using
python 3.4.5(cf. What’s New In Python 3.0) - scipy==0.18.0, numpy==1.9.0, matplotlib==1.5.2, sklearn==0.18.1, seaborn==0.7.1, pandas==0.18.1
$ pip3 install --user [package]install packages under~/.local- install jupyter to use ipython notebook
- Spell checking in Jupyter notebooks, Emacs keybindings for Jupyter notebook
Disclaimer
- At your own risk.
- This is NOT a complete list of options. See manual for it.
General Resources
- Python 3.4.3 documentation
- Scipy Lecture Notes
- NumPy Reference
- Introduction to machine learning with scikit-learn
Template
#!/usr/bin/python3 -tt
import numpy as np
import scipy.stats as stats
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
# The following lines are only for Jupyter
%matplotlib inline
from pylab import rcParams
rcParams['figure.figsize'] = 10, 5 ## width, height (inches)
Python Basics
Set Types
A constructor set() takes an iterable object.
Manual.
A = set(range(3,7)) # {3, 4, 5, 6}
- Propositions:
a in A,a not in A,A.issubset(B),A.issuperset(B) - Methods for tweak:
add(x),discard(x) - Binary operations:
A.intersection(B),A.union(B),A.difference(B)
Built-in modules
- itertools:
product(),permutations(),combinations(),combinations_with_replacement() - collections:
Counter(),defaultdict()
Datetime
A datetime object represents a date and a time. Manual.
from datetime import datetime
from pytz import timezone
now_de = datetime.now(timezone('Europe/Berlin')) # datetime.datetime object
now_jp = now_de.astimezone(timezone('Asia/Tokyo')) # in a different TZ
Use datetime.strptime() to convert a string into a datetime object.
datetime.strptime('2009/3/25 19:30:58', '%Y/%m/%d %H:%M:%S')
It is more practical to use pandas.to_datetime() because it automatically
guesses the format.
Manual.
pd.to_datetime('2009/3/25 19:30:58') # no need to give the format
The output is a pandas.tslib.Timestamp object, but we can regard it as a
datetime object.
.month,.day: return month and day, respectively.
To create a series of dates, pd.date_range is useful
pd.date_range('20160101', periods=2, freq='D')
# DatetimeIndex(['2016-01-01', '2016-01-02'], dtype='datetime64[ns]', freq='D')
Methods for a datetime object
date(): converts a datetime object into a date objectweekday(): gives the weekday in a number: 0 (mon) &ndash 6 (sun)isoformat(sep=' '): ISO 8601 format (for SQL)
A difference of times gives a timedelta
object. The .days attribute gives the time difference in days.
Files I/O
fo = open('test.txt','r') # creates a file object (for reading)
for line in fo: # reads the file line by line
line = line.rstrip() # removes white spaces
''' ... '''
fo.close # close the file object
If we use a with statement, we do not need to close the file object.
with open("test.txt") as fo:
for line in fo:
line = line.rstrip()
''' ... '''
- Manual: open(), rstrip(), file object, tutorial
fo.write("string")writes a string without "\n" in the writing mode.
JSON
- Manual
data = json.loads(json_string)json_string = json.dumps(data)
Use json.load(file_obj) (not loads()) to read a JSON file.
import json
json_fo = open("sample.json",'r') # the file object of the JSON file
data = json.load(json_fo) # read the file as a variable
json_fo.close()
Use json.dump(data,file_obj) (not dumps()) to create a JSON file.
jfo = open('write.json','w')
json.dump(data,jfo) # store the variable "data" as a json file
jfo.close()
SQLite3
Reads a sqlite3 database and execute something row by row. Manual, Tutorial.
import sqlite3
conn = sqlite3.connect('Auto.sqlite') # a connection object
#conn.row_factory = sqlite3.Row # for a dictionary cursor (*)
cur = conn.cursor() # a cursor object
cur.execute('SELECT * FROM Auto')
while True:
''' execute something row by row '''
row_tuple = cur.fetchone() # a row as a tuple (not a list!)
if row == None: # do not forget it
break
''' do something '''
conn.close()
cur.fetchone()returns a tuple of a row instead of a list. Uselist()to convert a tuple into a list.cur.fetchall()returns a list of tuples of all rows.- Using the line (*), we can create a dictionary cursor. After that
cur.fetchone()returns a sqlite Row object andcur.fetchall()returns a list of Row objects. A Row object behaves like a dictionary.
A placeholder works as follows:
conn = sqlite3.connect('Auto.sqlite') # a connection object
cur = conn.cursor() # a cursor object
cur.execute('UPDATE Auto SET name=? WHERE cylinders=?', ('something',4))
conn.commit()
conn.close()
- A placeholder prevents an SQL injection.
- If a placeholder is only one
?, then we have to use(something,)for a tuple. cur.executemany()accepts a tuple of tuples instead of a tuple.
Other Databases
- MySQL: mysqlclient, other possibility.
- MongoDB: PyMongo
- Hadoop Streaming: mrjob
- Spark: PySpark, MLlib
Regular Expression
import re
re.sub() corresponds s/regexp/alternative/g in Perl.
Manual.
re.sub('\d\d','---',"abc12def25g") # 'abc---def---g'
re.sub('\d\d',lambda x: str(int(x.group(0))**2),"abc12def25g") # 'abc144def625g'
re.search() corresponds /regexp/ in Perl and returns a match object.
match_obj = re.search('(\d\d).*(\d\d)','abc12de25')
match_obj.groups() # gives a tuple ('12', '25')
To get each matched string, use group(1), group(2), ... (Do not forget the
parentheses.)
Use the re.I flag for the case-insensitive match.
match_obj = re.search('AB','abc12de25', re.I) # match
re.findall()
re.findall() gives the list of matched strings:
re.findall('\d\d','abc12de25') # ['12', '25'] # (global match)
Mathematics and Statistics (numpy)
NumPy Reference. This section is basically part of An introduction to Numpy and Scipy.
Creating array objects
A (multi-dimensional) matrix can be represented as an array object (a.k.a. ndarray). The easiest ways to create one is to give a list to the np.array() function. Its size can be changed by reshape() method.
import numpy as np
np.array([[1,2,3],[4,5,6]]) # 2x3 matrix
# [[1 2 3]
# [4 5 6]]
np.array(range(1,7)).reshape((2,3)) # the same matrix as above
np.arange(1,7).reshape((2,3)) # this is also the same
To make a copy of an array object, use the copy() method.
np.arange(4): same asnp.array(range(4)).np.zeros((2,3)): the 2x3 matrix consisting only of 0np.ones((2,3)): the 2x3 matrix consisting only of 1np.identity(2): the 2x2 identity matrixnp.diag([2,3]): $\mathrm{diag}(2,3)$np.linspace(0,1,num=100): divides the closed interval $[0,1]$ with 100 points
Manipulation and slices
Use the shape property to find the size of an array object.
a = np.zeros((3,4))
a.shape # tuple (3,4)
a.shape[0] # 3
np.arange(2).shape # (2,)
The last array is a 1-dimentional array. Use np.newaxis to make it a row/column vector (or the reshape() method):
a = np.arange(3) # 1-dim array (3,)
a[:,np.newaxis] # column vector (3,1)
a[np.newaxis,:] # row vector (1,3)
We can slice part of an array as follows. (We should be careful about dimension.)
a = np.array([[1,2,3],[4,5,6]])
a[0,0] # 1
a[1,:] # the 2nd row [4,5,6] (1-dim array!)
a[:,2] # the 3rd column [3,6] (1-dim array!)
a[:,0:2] # the following 2x2 matrix
# [[1, 2]
# [4, 5]]
a[:,0:2] = np.identity(2) # A slice accepts a substitution
# [[1, 0, 3]
# [0, 1, 6]]
The np.concatenate() function concatenates two arrays
a = np.arange(2)[np.newaxis,:] # 1x2 matrix (row vector)
b = np.array([99,100])[np.newaxis,:] # 1x2 matrix (row vector)
np.concatenate((a,b),axis=0) # rbind()
# [[ 0 1] # (1+1)x2 matrix
# [ 99 100]]
np.concatenate((a,b),axis=1) # cbind()
# [[ 0 1 99 100] ] # 1x(2+2) matrix
Linear algebra
np.pi and np.e give the mathematical constants. A lot of mathematical functions such as np.abs(), np.sqrt(), np.log() are available. The np.sign() function returns the sign of an element as 1, 0 or -1.
np.sign(np.arange(-2,3)) # [-1, -1, 0, 1, 1]
The coordinate-wise binary operations: +, -, *, /, % (reminder). If the shapes of two matrices do not agree, then smaller one will be broadcasted. For example,
a = np.arange(1,7).reshape((2,3)) # 2x3 matrix
# [[1 2 3]
# [4 5 6]]
c = (np.arange(3)-1)[np.newaxis] # 1x3 matrix
# [[-1, 0, 1] ]
a+c
# [[0, 2, 4] # '+c' is applied to all rows
# [3, 5, 7]]
d = np.array([10,-10]).reshape((2,1)) # 2x1 matrix
# [[ 10]
# [-10]]
a*d
# [[ 10 20 30] # '*d' is applied to all columns
# [-60 -50 -40]]
If the larger matrix is square, then it is ambiguous how the array will be broadcasted. So it is a best practice to specify its shape by np.newaxis or reshape().
A multiplication of two matrices can be calculated by np.dot(). As its name suggests, it computes the dot product of two vectors (1-dim arrays).
np.dot(np.arange(3),np.array([0,3,10])) # 23
np.cross(u,v): the cross product $\vec u \times \vec v$ of two vectorsnp.outer(np.arange(1,10),np.arange(1,10)): multiplication tableX.tranpose(): $X^T$.
numpy.linalg
numpy.linalg contains lots of functions for linear algebra.
np.linalg.matrix_power(A,8): $A^8$np.linalg.inv(A): $A^{-1}$np.trace(A): $\mathrm{trace}(A)$. This is NOT in linalg.np.linalg.det(A): $\det A$np.linalg.matrix_rank(A): $\mathrm{rank}(A)$. This makes use of SVD.np.linalg.norm(A): Frobenius norm $\sqrt{\mathrm{trace}(A^TA)}$. More norms are available.
np.linalg.eig() computes eigenvalues and eigenvectors.
X = np.array([1,2,3,2,4,5,3,5,6]).reshape((3,3))
# [[1, 2, 3]
# [2, 4, 5]
# [3, 5, 6]]
evals, evecs = np.linalg.eig(X)
evals # eivenvalues (1-dim array)
# [ 11.34481428, -0.51572947, 0.17091519]
evecs # evecs[:,i] is an evals[i]-eigenvector.
# [[-0.32798528, -0.73697623, 0.59100905]
# [-0.59100905, -0.32798528, -0.73697623]
# [-0.73697623, 0.59100905, 0.32798528]]
np.dot(vecs, np.dot(np.diag(vals), np.linalg.inv(vecs))) # = X
The np.linalg.svd() function computes the singular value decomposition. Manual.
X = np.arange(15).reshape((3,5))
# [[ 0, 1, 2, 3, 4]
# [ 5, 6, 7, 8, 9]
# [10, 11, 12, 13, 14]]
U, s, Vt = np.linalg.svd(X)
S = np.diag(s) # makes s a diagonal matrix
np.dot(U, np.dot(S, Vt[0:len(s),:])) # very close to X
The actual size of U and Vt depend on the shape of X, so let us assume that $X$ is an $n \times p$ matrix with $n \leq p$. Then the SVD of $X$ is a decomposition $X=USV^T$ such that
- $S$ : the root of the $n \times n$ diagonal matrix of eigenvalues of the symmetric matrix $XX^T$.
- $U \in O(n)$ : the matrix the eigenvectors $XX^T$.
- $V$ : a $p \times n$ matrix whose columns are principal components of $X$. This is orthonormal in the sense that $V^TV = 1$.
The matrix Vt which np.linalg.sdv() gives $\tilde V^T$, where $\tilde V \in O(p)$ is an orthogonal extension of $V$. (Namely the orthonormal basis of $\ker(X)$ are added.)
Aggregation functions
Aggregation methods: sum(), prod(), mean(), var(), std(), max(), min().
Add the axis option to apply one of the methods along an axis.
X = np.array([-7,2,3,5,1,7,-4,8]).reshape((2,4))
# [[-7, 2, 3, 5]
# [ 1, 7, -4, 8]]
X.sum(axis=0) # sum() along the first axis
# [-6, 9, -1, 13] # 1-dim array
X.max(axis=1) # max() along the second axis
# [5, 8] # 1-dim array
Note that np.median() is a function.
Let $x^1$, ... , $x^n$ be row vectors of a matrix $X$. np.cov() computes the covariance of these vectors (i.e. the covariance matrix).
np.cov(X) # X is the 2x4 matrix as above.
# [[ 28.25 , 8.66666667]
# [ 8.66666667, 31.33333333]]
The (i,j)-component of np.cov(X) is $\frac{1}{n-1}\langle x^i_c, x^j_c\rangle$, where $x^i_c$ is the centred vector of $x^i$.
The correlations of rows can be computed by the np.corrcoef() function.
np.corrcoef(X) # X is the 2x4 matrix as above.
# [[ 1. , 0.29129939]
# [ 0.29129939, 1. ]]
Its $(i,j)$-component is equal to $\frac{1}{n-1}\langle x^i_n, x^j_n\rangle$, where $x^i_n$ is the normalised vector of $x^i_c$.
Use seaborn to draw a heat map of the correlation matrix: sns.heatmap(np.corrcoef(X)).
Random sampling (numpy.random)
- Manual.
np.random.seed(2016)sets a random seednp.random.choice(array, size=None, replace=True)returns random samples from the given array.
Statistical functions (scipy.stats)
scipy.stats consists of many functions for statistics.
Well-known distributions
Normal distribution
The PDF of $\mathcal N(\mu,\sigma)$ is given by $p(x) = \dfrac{1}{\sqrt{2\pi\sigma^2}}\exp\left(-\dfrac{(x-\mu)^2}{2\sigma^2}\right)$.
stats.norm.rvs(loc=0,scale=1,size=50): draws random samplesstats.norm.pdf(x,loc=0,scale=1): probability density functionstats.norm.cdf(x,loc=0,scale=1): cumulative density functionstats.norm.ppf(x,loc=0,scale=1): percent point function (inverse of CDF)- A multivariate normal distribution $\mathcal N(\mu,\Sigma)$ is also available.
Replacing norm with other term, we can use other continuous distribution
in a similar way.
Uniform distribution
The PDF of $\mathsf U(a,b)$ is given by $p(x) = \dfrac{1_{[a,b]}(x)}{b-a}$.
stats.uniform.rvs(loc=0,scale=1,size=50): draws random samplesstats.uniform.pdf(x,loc=0,scale=1): PDFlocmeans $a$ andscalemeans $b-a$.
Exponential distribution
The PDF of $\mathsf{Exp}(\lambda)$ is given by $p(x) = \lambda e^{-\lambda x}1_{[0,\infty)}$.
stats.expon.rvs(scale=1, size=50): draws random samplesstats.expon.pdf(x,scale=1): PDFscalemeans $1/\lambda$, which equals to the standard deviation.
Binomial distribution
The PMF of $\mathsf{Bin}(n,p)$ is given by $\mathbb P(X=x) = \begin{pmatrix} n \\ x\end{pmatrix} p^x(1-p)^{n-x}$ for $x = 0, \cdots, n$.
stats.binom.rvs(n,p,size=50): draws random samplesstats.binom.pmf(x,n,p): PMF (not PDF)stats.binom.cdf(x,n,p): CDFstats.binom.ppf(x,n,p): percent point function (the "inverse" of CDF)
Replacing binom with other term, we can use other discrete distribution in a similar way.
Possion distribution
The PMF of $\mathsf{Poisson}(\mu)$ is given by $\mathbb P(X=k) = \dfrac{\mu^k}{k!}e^{-\mu}$ for $k = 0, 1, 2, \cdots$.
stats.poisson.rvs(mu, size=1)stats.poisson.pmf(k, mu): PMF
Welch's t-test
- Assumptions: two random variables are normally distributed.
- Null Hypothesis: two random variables has the same expected value.
scipy.stats.ttest_ind() computes the p-value. Manual.
a = st.norm.rvs(loc=11,scale=5,size=200) # population A
b = st.norm.rvs(loc=10,scale=5.2,size=200) # population B
tstat, pval = st.ttest_ind(a, b, equal_var=False)
Fisher's exact test
- Assumptions: two random variables are Bernoulli distributed.
- Null Hypothesis: two random variables has the same expected value.
scipy.stats.fisher_exact() computes the p-value.
Manual.
But we have to calculate a crosstab in advance.
a = st.bernoulli.rvs(0.03, size=1200) ## population A
b = st.bernoulli.rvs(0.035, size=1000) ## population B
pos = np.array([a.sum(), b.sum()]) ## 1st row
neg = np.array([1200, 1000]) - pos ## 2nd row
ctab = np.array([neg,pos]) ## 2x2 table
oddsratio, pval = st.stats.fisher_exact(ctab)
A chi-squared test for goodness of fit
- Assumptions: two random variables are multinomial distributed.
- Null Hypothesis: two random variables are independent.
scipy.stats.chi2_contingency() computes the p-value. Manual.
We have to compute the crosstab in advance. (pandas.crosstab() is
convenient for the computation.)
a = ['abc'[x] for x in st.randint.rvs(0,3,size=300)] ## variable 1
b = ['yn'[x] for x in st.bernoulli.rvs(0.35, size=300)] ## variable 2
ctab = pd.crosstab(pd.Series(a, name='A'), pd.Series(b, name='B'))
chi2, pval, dof, expected = st.chi2_contingency(ctab)
Other useful modules
scipy.special(Manual).lbeta, etc.scipy.optimize(Manual). for gradient descent, etc. Note that the argument of the objective function must be an array.
DataFrame (pandas)
- pandas : the official website
- Intro to pandas data structures, An introduction to Pandas : tutorials
Note that we need to use print() to print the actual data in the following codes (not on the IPython).
Series
A Series is like list with keys (index). We can create one from an usual list as follows.
x = pd.Series(['a','b','c'], index=range(3))
x
# 0 a
# 1 b
# 2 c
# dtype: object
An object of Series behaves like a list in R.
x == 'a'
# 0 True
# 1 False
# 2 False
# dtype: bool
x[x=='a']
0 a
dtype: object
Examples of methods mean(), unique(), nunique(), value_counts(), etc.
np.random.seed(0)
x = pd.Series(np.random.randint(0,4,size=10))
x.unique() ## array([0, 3, 1])
x.nunique() ## len(x.unique()) i.e. 3
x.value_counts()
# 3 6
# 1 2
# 0 2
# dtype: int64
DataFrame
Something like a data frame in R.
data = {
'col1' : range(1,9),
'col2' : [x**2 for x in range(1,9)],
'col3' : [np.exp(x) for x in range(1,9)],
}
df = pd.DataFrame(data,columns=['col1','col2','col3']) # dict -> data frame
CSV
The method read_csv() is used to read a CSV file
df = pd.read_csv("file",sep=',')
file: a file location or a URL of the CSVindex_col=None: the column number of indexesheader=None, names=cols: If the file contains no header, we give names to the columns withnames(notcolumns)na_values='?': specified string is recognised as a NaNdecimal='.': decimal pointthousands=None: thousands separatordtype={'age':np.int64}: specified the type of a columnconverters={'date':pd.to_datetime}: apply a function to values in a column
To save an object of DataFrame, use df.to_csv("output.csv",index=False).
SQLite3
See the documentation for more details..
from pandas.io import sql
import sqlite3
conn = sqlite3.connect('db.sqlite')
query = "SELECT rowid,* FROM Table;"
df = sql.read_sql(query, con=conn)
See the data frame
url = "http://www-bcf.usc.edu/~gareth/ISL/Auto.csv"
df = pd.read_csv(url, na_values='?')
df.shape: tuple of the numbers of rows and columnsdf.info(): check the types of columns (find columns in "object")
df.describe() is something like summary(df) in R. (Note that the column of "object" is ignored):
mpg cylinders displacement weight acceleration \
count 397.000000 397.000000 397.000000 397.000000 397.000000
mean 23.515869 5.458438 193.532746 2970.261965 15.555668
std 7.825804 1.701577 104.379583 847.904119 2.749995
min 9.000000 3.000000 68.000000 1613.000000 8.000000
25% 17.500000 4.000000 104.000000 2223.000000 13.800000
50% 23.000000 4.000000 146.000000 2800.000000 15.500000
75% 29.000000 8.000000 262.000000 3609.000000 17.100000
max 46.600000 8.000000 455.000000 5140.000000 24.800000
year origin
count 397.000000 397.000000
mean 75.994962 1.574307
std 3.690005 0.802549
min 70.000000 1.000000
25% 73.000000 1.000000
50% 76.000000 1.000000
75% 79.000000 2.000000
max 82.000000 3.000000
Rows and columns
The "index" of rows (resp. columns) start from 0.
df.columns: list of the column namesdf.index: list of the row indexesdf.values: convertdfinto an ndarray object of numpy
The column named weight can be access by df['weight'] or df.weight.
0 3504
1 3693
...
395 2625
396 2720
Name: weight, dtype: int64
If "values" is used as a label of a column, df.values gives the result of the method values. Because many methods are defined on a data frame, we should NOT use df.col_name to get a column series.
We may change a label of a column by rename method.
df.rename(columns={'horsepower':'pow'},inplace=True) ## 'pow' is also a method...
Or we can assign a complete list of labels:
df.columns = ['mpg','cyl','disp','pow','weight','acc','year','origin','name']
df.index = ["r%s" % i for i in range(1,df.shape[0]+1)] ### We start with "r1"!
df.set_index('column_name',inplace=True) makes to make a column the row indexes.
While df['cyl'] gives the Series of the column "cyl", df.ix["r100"] gives the Series of the row "r100".
mpg 18
cyl 6
disp 232
pow 100
weight 2945
acc 16
year 73
origin 1
name amc hornet
Name: r100, dtype: object
Then df.ix["r100","weight"] gives the 2945.
df.ix accepts the notation :. For example df.ix["r100":"r103","cyl":"year"] gives
cyl disp pow weight acc year
r100 6 232 100 2945 16.0 73
r101 6 250 88 3021 16.5 73
r102 6 198 95 2904 16.0 73
r103 4 97 46 1950 21.0 73
To specify all columns (or rows) we may leave only :. For example df.ix[99:102,:] gives
mpg cyl disp pow weight acc year origin name
r100 18 6 232 100 2945 16.0 73 1 amc hornet
r101 18 6 250 88 3021 16.5 73 1 ford maverick
r102 23 6 198 95 2904 16.0 73 1 plymouth duster
Note that df.index[99] is "r100" and df.index[102] is "r103". (We have assigned r1 to df.index[0].) Namely the behaviour of : depends which we use to specify rows (or columns): labels or coordinates.
Remark. There are other ways to get values by using coordinates or labels.
The function pd.crosstab produces a contingency table.
pd.crosstab(df['cyl'],df['year'])
# year 70 71 72 73 74 75 76 77 78 79 80 81 82
# cyl
# 3 0 0 1 1 0 0 0 1 0 0 1 0 0
# 4 7 13 14 11 15 12 15 14 17 12 25 21 27
# 5 0 0 0 0 0 0 0 0 1 1 1 0 0
# 6 4 8 0 8 7 12 10 5 12 6 2 7 3
# 8 18 7 13 20 5 6 9 8 6 10 0 1 0
Conversion of a data type
Convert an "object" to a "float".
df['pow'].dtype # dtype: object
df['pow'] = pd.to_numeric(df['pow'],errors='coerce') # dtype: float64
Convert a date string (object) to a datetime object.
s = pd.Series(['12/1/2012', '30/01/2012']) # dtype: object
s = pd.to_datetime(s, format='%d/%m/%Y') # dtype: datetime[64]
to_datetime() can guess the format of the date automatically. So we leave out the format option, but the process will be faster if the format is given.
Convert a column to a categorical data.
df['origin'].unique() # [1 3 2] # dtype: int64
df['origin'] = df['origin'].astype('category') # dtype: category
df['name'].dtype # dtype: object
df['name'] = df['name'].astype('category') # dtype: category
The following code makes the categories more meaningful.
df['origin'].cat.categories = ['usa','europe','japan']
Concatenate dataframes
pd.concat([df,dg]): equivalent torbind(df,dg)in R.pd.concat([df,dg],axis=1): equivalent tocbind(df,dg)in R.
Merge
data1 = {'rowid' : range(1,4), 'col1' : range(10,13),}
df = pd.DataFrame(data1,columns=['rowid','col1'])
data2 = {'rowid' : range(2,5), 'col2' : ['a','b','c'],}
dg = pd.DataFrame(data2,columns=['rowid','col2'])
pd.merge(df,dg,on='rowid',how='inner') merge two dataframes for only common rowids.
rowid col1 col2
0 2 11 a
1 3 12 b
how='left' (resp. how='right') option keeps all rows of the first (resp. second) dataframe.
rowid col1 col2
0 1 10 NaN
1 2 11 a
2 3 12 b
how='outer' option keeps all rows of the both dataframes.
Applying a function to a column/row
To apply a function to each column, use apply method.
dg = df.ix[:,'mpg':'acc'] # consists only of numerical columns.
dg.apply(np.median,axis=0) # applying an aggregate function
# mpg 23.0
# cyl 4.0
# disp 146.0
# pow 95.0
# weight 2800.0
# acc 15.5
# dtype: float64
dg.apply(lambda x: np.log(x+1)).head(3) # applying a component-wise function
# mpg cyl disp pow weight acc
# r1 2.944439 2.197225 5.730100 4.875197 8.161946 2.564949
# r2 2.772589 2.197225 5.860786 5.111988 8.214465 2.525729
# r3 2.944439 2.197225 5.765191 5.017280 8.142354 2.484907
We may also use apply method for a series, when we want to apply a component-wise function.
df['disp'].apply(lambda y: np.log(y+1)).head(3)
# r1 5.730100
# r2 5.860786
# r3 5.765191
# Name: disp, dtype: float64
If we want to apply an aggregate function (of numpy) to a series, use it directly.
np.median(df['disp']) # 146.0
To apply a function to each row, use axis=1 option.
Missing data
pd.isnull(df)gives a boolean data frame giving "True" where "NaN" was there.df.dropna(how='any')gives only rows without "NaN".df.fillna(value=0)fills "0" at missing points.
To count the number of NA by column:
from collections import Counter
pd.isnull(df).apply(lambda x: Counter(x)[True])
# mpg 0
# cyl 0
# disp 0
# pow 5
# weight 0
# acc 0
# year 0
# origin 0
# name 0
# dtype: int64
To see the rows with a missing value, we make a boolean series as follows.
from collections import Counter
row_with_na = pd.isnull(df).apply(lambda x: Counter(x)[True]>0,axis=1)
df[row_with_na]
# mpg cyl disp pow weight acc year origin name
# r33 25.0 4 98 NaN 2046 19.0 71 1 ford pinto
# r127 21.0 6 200 NaN 2875 17.0 74 1 ford maverick
# r331 40.9 4 85 NaN 1835 17.3 80 2 renault lecar deluxe
# r337 23.6 4 140 NaN 2905 14.3 80 1 ford mustang cobra
# r355 34.5 4 100 NaN 2320 15.8 81 2 renault 18i
From a wide table to a long one, and back
We use the following wide data frame.
dg = df[['mpg','disp','pow','name']].head(2)
# mpg disp pow name
# r1 18 307 130 chevrolet chevelle malibu
# r2 15 350 165 buick skylark 320
The pd.melt() method makes a wide data frame into a long one.
dg.reset_index(inplace=True) # row indexes -> a column
dg_long = pd.melt(dg,id_vars=['index'])
index variable value
# 0 r1 mpg 18
# 1 r2 mpg 15
# 2 r1 disp 307
# 3 r2 disp 350
# 4 r1 pow 130
# 5 r2 pow 165
# 6 r1 name chevrolet chevelle malibu
# 7 r2 name buick skylark 320
The pivot() method makes a long data frame into a wide one.
dg_wide = dg_long.pivot(index='index',columns='variable',values='value')
# variable disp mpg name pow
# index
# r1 307 18 chevrolet chevelle malibu 130
# r2 350 15 buick skylark 320 165
When the data table has duplicate rows, we should use pivot_table().
Functions corresponding to dplyr
Warning. One of the important properties of dplyr of R is: the output is always a new data frame. But I have not checked whether (most of) the following codes produce new data frame or not. If not, we might want to use the copy method.
filtering rows
If s is a boolean series, then df[s] consists of rows where "s" is True.
df[ (df['mpg'] == 18) & (df['acc'] <= 12) ] # Don't forget the round brackets!
# mpg cyl disp pow weight acc year origin name
# r1 18 8 307 130 3504 12 70 1 chevrolet chevelle malibu
# r3 18 8 318 150 3436 11 70 1 plymouth satellite
The isin() method may be useful.
df[ df['name'].isin(['honda civic cvcc','toyota corolla 1200']) ]
# mpg cyl disp pow weight acc year origin name
# r54 31.0 4 71 65 1773 19.0 71 3 toyota corolla 1200
# r132 32.0 4 71 65 1836 21.0 74 3 toyota corolla 1200
# r182 33.0 4 91 53 1795 17.5 75 3 honda civic cvcc
# r249 36.1 4 91 60 1800 16.4 78 3 honda civic cvcc
When using a regular expression, we should use bool function to get a boolean series.
df[ df['name'].apply(lambda x: bool(re.search('^h.*c$',x))) ]
# mpg cyl disp pow weight acc year origin name
# r150 24.0 4 120 97 2489 15.0 74 3 honda civic
# r182 33.0 4 91 53 1795 17.5 75 3 honda civic cvcc
# r199 33.0 4 91 53 1795 17.4 76 3 honda civic
# r217 31.5 4 98 68 2045 18.5 77 3 honda accord cvcc
# r249 36.1 4 91 60 1800 16.4 78 3 honda civic cvcc
# r383 38.0 4 91 67 1965 15.0 82 3 honda civic
arrange rows
df.sort_values(by="disp",ascending=True) sorts rows by values of "disp". The option ascending is "True" by default. The option by accepts a list as well.
df.sort_values(by=["disp","pow"])).head(5)
# mpg cyl disp pow weight acc year origin name
# r118 29.0 4 68 49 1867 19.5 73 2 fiat 128
# r112 18.0 3 70 90 2124 13.5 73 3 maxda rx3
# r72 19.0 3 70 97 2330 13.5 72 3 mazda rx2 coupe
# r335 23.7 3 70 100 2420 12.5 80 3 mazda rx-7 gs
# r54 31.0 4 71 65 1773 19.0 71 3 toyota corolla 1200
select columns
If s is a list of column names, then df[s] gives a data frame consisting of columns in s.
cols = ['pow','mpg','cyl']
df[cols].head(3)
# pow mpg cyl
# r1 130 18 8
# r2 165 15 8
# r3 150 18 8
Use the drop(cols, axis=1) method to remove a few columns. Manual.
mutate
Unfortunately I have not found any standard way to achieve mutate of R in Python. Because many operation such as "+" can be applied to series by coordinates,
df['new_col'] = 3*df['mpg']**2 - 4*df['pow']
adds a column "new_col" to "df". Or we may use apply(func,axis=0) to apply a function taking a series of a row.
df['new_col'] = df.apply(lambda x: 3*x['mpg']**2-4*x['pow'], axis=1)
When we need a new data frame, use df_new = df.copy() to duplicate a data frame.
Grouping
groupby('col_name') method splits a data frame by values of 'col_name' so that we can deal with them at the same time. (Manual)
df['origin'].unique()
# [1, 3, 2]
# Categories (3, int64): [1, 3, 2]
df.groupby('origin').mean()
# mpg cyl disp pow weight acc \
# origin
# 1 20.071774 6.258065 246.284274 119.048980 3363.250000 15.011694
# 2 27.891429 4.157143 109.142857 80.558824 2423.300000 16.787143
# 3 30.450633 4.101266 102.708861 79.835443 2221.227848 16.172152
#
# year
# origin
# 1 75.584677
# 2 75.814286
# 3 77.443038
The method groupby accepts a list of column names as well.
Visualisation (matplotlib, seaborn)
- matplotlib, Gallery, Pyplot tutorial, User's Guide (including Beginner's Guide)
- Seaborn, API, Gallery
- Visualisation through pandas
- Python data visualizations on the Iris dataset
The following code produces a 400x400 image "graph.png".
plt.figure(figsize=(400/96,400/96),dpi=96)
''' plot something ... '''
plt.tight_layout() # when producing a small image
plt.savefig('graph.png',dpi=96)
Using plt.show() instead plt.savefig(), we obtain an image window.
Note that need %matplotlib inline to show the image on IPython Notebook.
Simple diagrams
- A basic strategy is: use
pandasorseabornif possible. pandasoften requires a wide table. Usepivot(columns=, values=)(with or withoutindexoption).- Some methods raises an error if there is a missing value in a data.
Use
dropna(how='any')in such a case.
Line chart
df_tmp = df.groupby(['year','origin'])['horsepower']\
.agg({'avg':np.mean}).reset_index()
df_tmp.pivot(index='year',columns='origin',values='avg').plot()
plt.ylabel('mean(horsepower)')
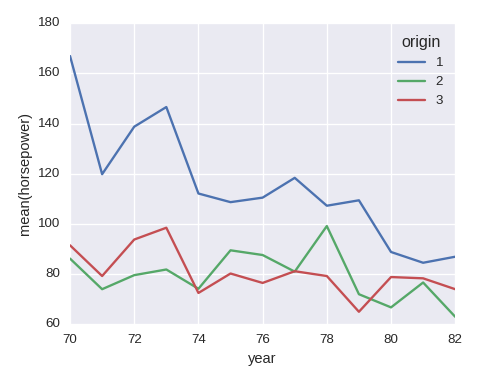
Histogram
Put stacked=True in plt.hist() if a stacked histgram is needed.
yvals = sorted(df.origin.unique())
data = [df.weight[df.origin==y].dropna(how='any') for y in yvals]
plt.hist(data,bins=20,label=yvals)
plt.xlabel('weight')
plt.ylabel('count')
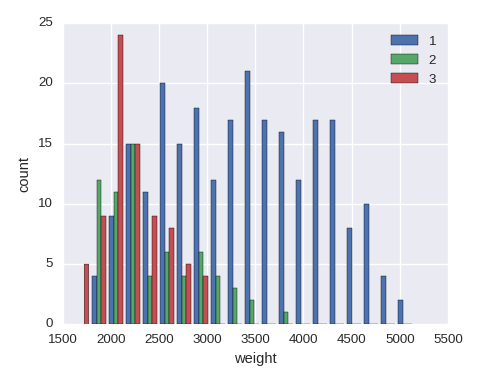
The simplest histogram can be obtained df.weight.hist().
Box plot
sns.boxplot(x='origin',y='horsepower',data=df,orient='v')
sns.swarmplot(x='origin',y='horsepower',data=df,orient='v',alpha=0.2)
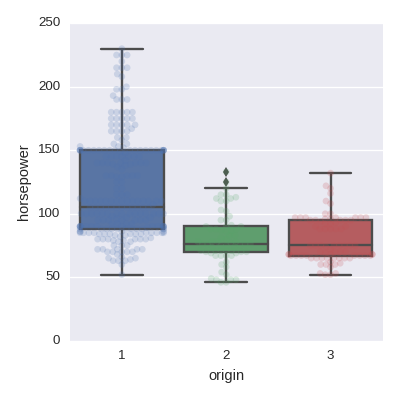
Scatter plot
df.plot.scatter(x='horsepower',y='weight',alpha=0.5)
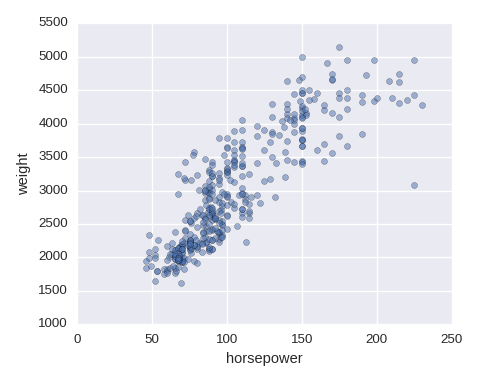
To add a color from a categorical variable, use FacetGrid. size=5 is for
the size of the image
num1,num2 = 'horsepower', 'weight'
sns.FacetGrid(df,hue="origin",size=5)\
.map(plt.scatter,num1,num2,alpha=0.5).add_legend()
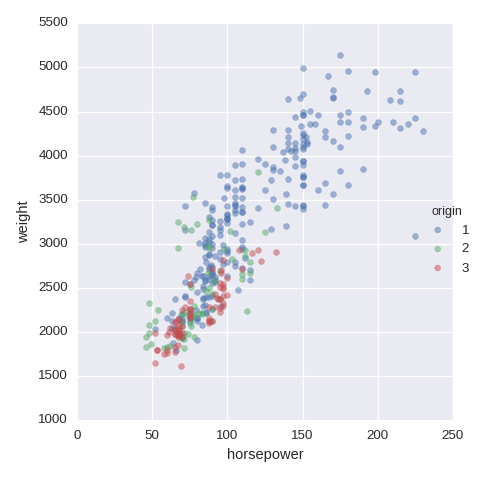
Examples of visualisation
A numerical variable vs a categorical variable
sns.boxplot(x='horsepower',y='origin',data=df,orient='h')
sns.swarmplot(x='horsepower',y='origin',data=df,orient='h',color='0.2',alpha=0.2)
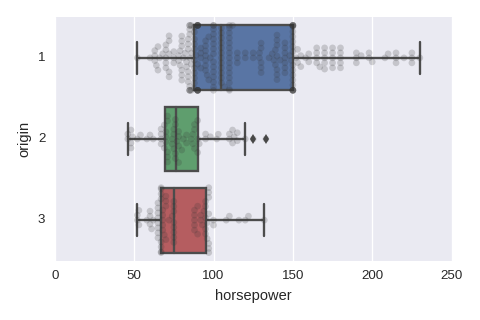
Numerical variables vs a categorical variable
To adjust the size of the image put figsize=(480/96,420/96) in plt.subplots().
cols = ['horsepower','displacement','weight']
fig, axes = plt.subplots(nrows=len(cols),ncols=1)
for col,subax in zip(cols,list(axes.flat)):
sns.boxplot(x=col,y='origin',data=df,orient='h',ax=subax)
sns.swarmplot(x=col,y='origin',data=df,orient='h',alpha=0.2,ax=subax)
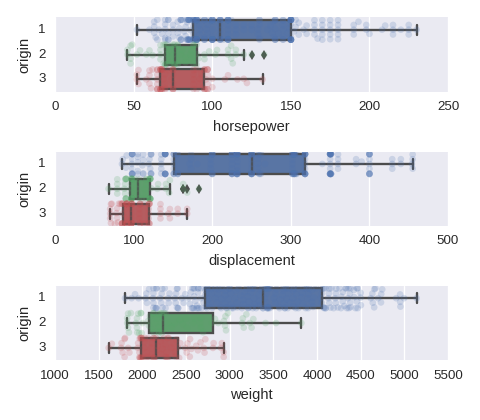
A categorical variable vs a categorical variable
The proportions of the target values in each value. (The size is not adjustable.)
cat1,cat2 = 'cylinders','origin' # cat vs cat
df_gb0 = df.groupby([cat1,cat2]).size().rename('cnt').reset_index()
df_gb1 = df_gb0.groupby(cat1)['cnt'].agg({'size':np.sum}).reset_index()
df_tmp = pd.merge(df_gb0,df_gb1,on=cat1)
df_tmp['rate'] = df_tmp['cnt']/df_tmp['size']
df_tmp = df_tmp.pivot(index=cat1,columns=cat2,values='rate').fillna(0)
df_tmp.plot.bar(stacked=True)
plt.legend(bbox_to_anchor=(1.1, 1),title=cat1)
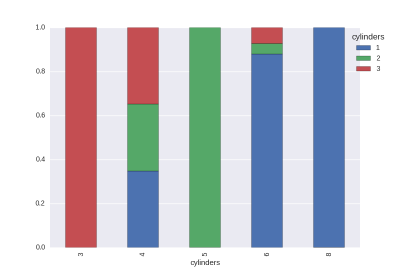
The histogram is easy to draw.
sns.countplot(x='cylinders',hue='origin',data=df)
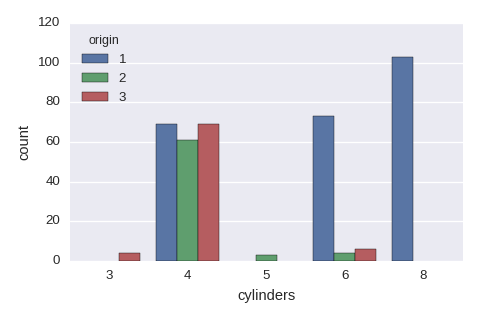
Categorical variables vs a categorical variable
df.mpg = pd.cut(df.mpg,bins=4) ## a new cat variable
cols = ['cylinders','mpg']
fig, axes = plt.subplots(nrows=len(cols),ncols=1,figsize=(480/96,420/96))
for col,subax in zip(cols,list(axes.flat)):
sns.countplot(x=col,hue='origin',data=df,ax=subax)
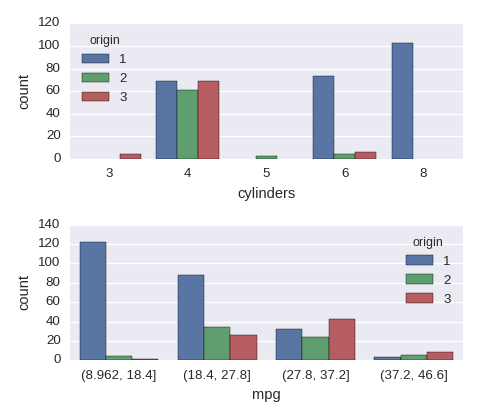
A numerical variable vs a numerical variable
See the section "scatter plot" above.
Numerical variables vs a numerical variable
If there are too many ticks on the y-axis, then use mathplotlib.ticker.MultipleLocator.
import matplotlib.ticker as ticker ## if needed
cols = ['horsepower','displacement','acceleration']
fig, axes = plt.subplots(nrows=len(cols),ncols=1,figsize=(480/96,480/96))
for col,subax in zip(cols,list(axes.flat)):
df.plot.scatter(x=col,y='weight',alpha=0.5,ax=subax)
subax.yaxis.set_major_locator(ticker.MultipleLocator(1000)) # if needed
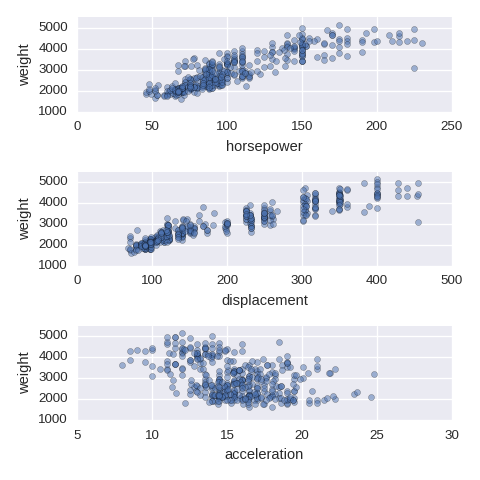
A categorical variable vs a numerical varaible
See section "Box plot" above.
Categorical variables vs a numerical varaible
cols = ['origin','cylinders']
fig, axes = plt.subplots(nrows=len(cols),ncols=1,figsize=(480/96,480/96))
for col,subax in zip(cols,list(axes.flat)):
sns.boxplot(x=col,y='horsepower',data=df,orient='v',ax=subax)
sns.swarmplot(x=col,y='horsepower',data=df,orient='v',alpha=0.2,ax=subax)
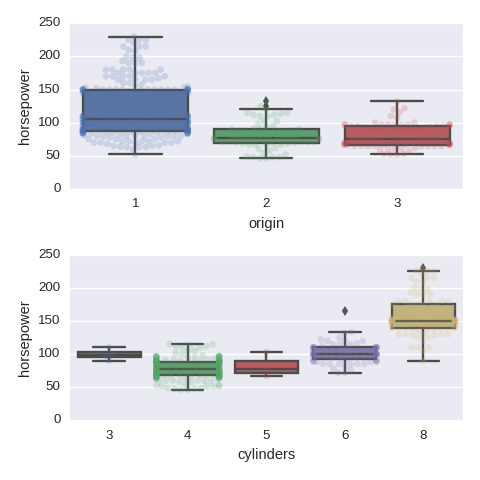
Pairwise relationships between numerical variables
sns.pairplot() shows pairwise relationships between (all) pairs of numerical
variables.
cols = ['mpg','displacement','acceleration','origin']
df_tmp = df[cols].copy()
df_tmp.origin = df_tmp.origin.map({1:'usa',2:'eur',3:'jpn'})
sns.pairplot(df_tmp,hue='origin',size=2)
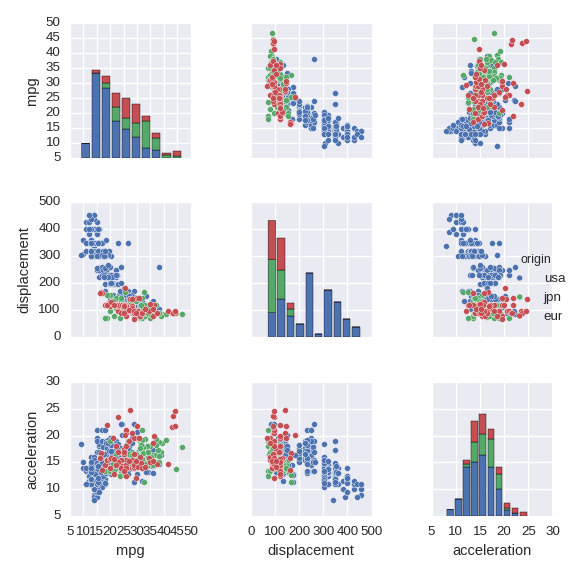
- For the
hueoption a categorical variable must be non numerical. Even a string of a number (such as '1') is not allowed. sizespecifies the size of the image.
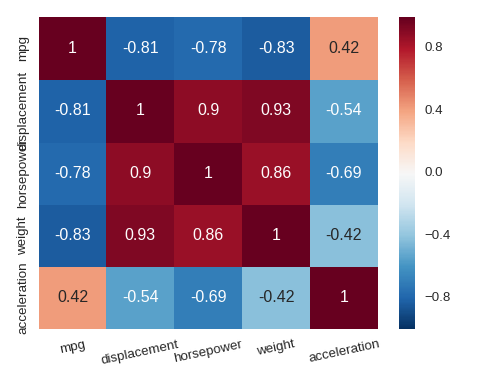
Heat map
sns.heatmap() can be used to visualise a matrix such as a correlation matrix.
cols = ['mpg','displacement','horsepower','weight','acceleration']
plt.xticks(rotation=12)
sns.heatmap(df[cols].corr(),annot=True)
Machine Learning (scikit-learn)
The feature matrix and the response vector are the NumPy arrays and stored on
different variables: X and y, respectively. They are allowed to have only
numerical values.
sample data
iris = load_iris()
print(iris.DESCR) # show the description of the dataset.
X = pd.DataFrame(iris.data,columns=iris.feature_names)
y = pd.Series(iris.target,name='class')
To obtain an array of objects (rather than the numbers) for the target
variable, we use iris.target_names:
[ iris.target_names[c] for c in iris.target ]
Train-Test split
from sklearn.model_selection import train_test_split
X_train,X_test,y_train,y_test = train_test_split(X,y,test_size=0.4,random_state=4)
random_state is something like seed().
Label Binalizer
We have to convert a categorical column into a pivot table. But we should use
LabelBinarizer to create a table instead of pandas pivot() for
consistency.
from sklearn.preprocessing import LabelBinarizer
lb = LabelBinarizer()
lb.fit(y)
pd.DataFrame(lb.transform(y), columns=lb.classes_) ## pivot table
The following function creates a function converting a categorical column (series) into a pivot table.
def pivot(vec,vals=None):
if vals is None:
vals = vec.unique()
lb = LabelBinarizer().fit(vec)
def create_df(series):
pivot_df = pd.DataFrame(lb.transform(series),columns=lb.classes_,index=series.index)
return pivot_df[vals]
return create_df
Dimension Reduction
Principal Component Analysis. Manual
from sklearn.decomposition import PCA
pca = PCA(n_components=2) # specify the number of principal components
X_pca = pca.fit_transform(X) # describe X by principal components
pca.explained_variance_ratio_ # the variance of each components (ratio)
t-SNE. Manual.
from sklearn.manifold import TSNE
sne = TSNE(n_components=2, random_state=2)
np.set_printoptions(suppress=True)
sne_xy = sne.fit_transform(X) ### each row gives a 2-dim coordinate
Resampling
from imblearn.over_sampling import SMOTE
sm = SMOTE(random_state=3)
X_train, y_train = sm.fit_sample(X_train, y_train)
The output (X_train and y_train) are both NumPy arrays.
Models
Linear Regression
from sklearn.linear_model import LinearRegression
Elastic Net
from sklearn.linear_model import ElasticNet
param_enet = {'alpha': [0.01,0.1,1], 'l1_ratio': [0.1,0.4,0.7,1]}
The cost function is defined by
$$J(\beta) = \frac{1}{2n}\|y-X\beta\|^2 + \alpha \left(\rho \|\beta'\|_1 + \frac{1-\rho}{2} \| \beta' \|_2^2 \right).$$
Here $n$ is the number of samples and $\beta' = (\beta_1,\cdots,\beta_p)$. (Namely the intercept is removed.)
- If $\rho=0$, then the model is a ridge regression.
- If $\rho=1$, then the model is a LASSO.
But
l1_ratiomust be > 0.01 because of the implementation. - $\alpha$ is a positive real number.
- Use
pd.Series(fit_enet.coef_,index=X.columns)to check the trained coefficients. - Manual, LASSO.
Logistic Regression
from sklearn.linear_model import LogisticRegression
param_enet = {'penalty': ['l1','l2'], 'C': [0.01,0.1,1,10,100]}
This is actually a penalised logistic regression (a.k.a. regularised logistic regression). The cost function (with $L_2$ penalty) is defined by
$$J(\beta) = C \sum_{i=1}^n \log\left(\exp(-y^i\langle x^i,\beta \rangle)+1\right) + \frac{1}{2}\|\beta'\|_2^2$$
- Manual.
penalty: the norm for the penalty term.l1orl2.C: $1/\lambda$ ($\lambda$ is called "regularization strength" in the manual.)class_weight: the weights of the classes.- If it is not given, then all classes have weight one.
{'class0':1, 'class1':2}specifies the weight of classes explicitlybalanced: adjusts weights inversely proportional to class frequencies
K-Nearest Neibourhood
from sklearn.neighbors import KNeighborsClassifier
param_knn = {'n_neighbors':[1,3,5]}
- Manual
KNeighborsRegressoris for a regression.
Decision Tree
from sklearn.tree from DecisionTreeClassifier
param_tree = {'max_leaf_nodes': [3,6,12,24,48]}
DecisionTreeRegressoris for a regression.random_state: a seed for a random numbermax_features: the number of features to look for the best split. The default value isNone(i.e. the all features).max_depth: the maximum depth of tree. This conflictsmax_leaf_nodesmax_leaf_nodes: grow a tree with a given number in best-first fashion. This conflictsmax_depth.- Manual
To see the importance of the features (after training).
pd.Series(estimator_tree.feature_importances_, index=X.columns)
Here we assume that X is a data frame of features.
To see the result we use tree.export_graphviz.
On Jupyter Notebook:
from sklearn.externals.six import StringIO
from sklearn.tree import export_graphviz
from IPython.display import Image
import pydot
dot_data = StringIO()
export_graphviz(clf, out_file=dot_data,
feature_names=list(X.columns),
#class_names=iris.target_names, ## for classification
filled=True, rounded=True, special_characters=True)
graph = pydot.graph_from_dot_data(dot_data.getvalue())[0]
Image(graph.create_png())
To create a PNG file for the tree:
from sklearn.tree import export_graphviz
with open("tree.dot", 'w') as f:
f = export_graphviz(grid_tree.best_estimator_, out_file=f,
filled=True, rounded=True, special_characters=True,
#class_names=iris.target_names, ## for classification
feature_names=list(X.columns))
Then we can convert the dot file "tree.dot" into a PNG file.
$ dot -Tpng tree.dot -o tree.png # shell command
Random Forest
from sklearn.ensemble import RandomForestClassifier
param_rf = {'n_estimators': [10,30], 'max_depth': [3,5,7,None]}
- Manual.
RandomForestRegressoris for a regression random_state(int) : a seed of random numbersn_estimators: the number of trees in the forestmax_depth: the maximum depth of the trees
We can see the importances of features in the same way as a decision tree.
pd.Series(estimator_rf.feature_importances_, index=X.columns)
Support Vector Machine
from sklearn.svm import SVC
param_svm = {'C': [0.01,0.1,1,10], 'kernel': ['rbf','sigmoid','linear']}
- SVC, SVR (for regression)
C (=1.0): penalty parameter of the error terms. (a.k.a the cost of SVC)kernel (='rbf'): kernel function. ‘linear’, ‘poly’, ‘rbf’, ‘sigmoid’, ‘precomputed’, etc.- $K(\vec x_1,\vec x_2) = \exp\bigl(-\gamma\| \vec x_1 - \vec x_2\|\bigr)$ : the radial kernel (rbf)
- $\tanh(\gamma\langle \vec x_1,\vec x_2 \rangle + r)$
: the sigmoid kernel ($r$ is specified by
coef0) - $(\gamma\langle \vec x_1,\vec x_2\rangle+r)^d$
: the polynomial kernel ($d$ is specified by
degree(=3))
gamma: $\gamma$ in the above kernels. Note that the default value is 0.verbose (=False)
XGBoosting
XGBoost is a boosting model of trees.
from xgboost import XGBClassifier
param_xgb = {
'learning_rate' : [0.01,0.1],
'n_estimators' : [5,10,20],
'max_depth' : [5,10],
'reg_alpha' : [0.01,0.1],
'reg_lambda' : [0.01,0.1]
}
The objective function for the t-th step is given by $$\mathrm{Obj}^{(t)} = \sum_{i=1}^n \left( g_i f_t(x_i) + \frac{1}{2}h_i f_t(x_i)^2 \right) + \gamma T + \dfrac{\lambda}{2} \|w\|^2.$$ Here $g_i$ and $h_i$ are 1st and 2nd derivatives of the given loss function, $f_t$ is a tree model to learn, $T$ is the number of leaves and $w = (w_1,\cdots,w_T)$ is scores of the tree. $\gamma$ and $\lambda$ are (nonnegative) tuning parameters of the model.
- mathematical model, API Reference, Code Examples, tuning parameter
XGBRegressoris for a regressionxgboostdoes not belong to scikit-learn. But it provides a wrapper class for scikit-learn.max_depth(=6) : maximum depth of a tree.gamma(=0) : regularization coefficient on the number of leaves. (See the objective function above.)reg_lambda(=1) : L2 regularization term on scores of a treereg_alpha(=0) : L1 regularization term on scores of a tree
Clustering
K-Means
from sklearn.cluster import KMeans
km = KMeans(n_clusters=8,n_init=10,init='random',verbose=0)
km.fit(X)
Manual. The result can be obtained by km.predict(X).
n_clusters: the number of clustersn_init: the number of trialsmax_iter: the maximum number of iterations (in a single trial)init: the way to choice the initial centroids:k-means++(default),randomor an ndarrayrandom_state: a random seedverbose: verbosity mode
Model Evaluation
K-Fold Cross-Validation
cross_val_score computes accuracy scores of trials of cross-validation.
from sklearn.cross_validation import cross_val_score
scores = cross_val_score(estimator, X, y, cv=10, scoring='accuracy')
print(scores.mean()) # an estimate of out-of-sample accuracy
estimator is the instance of an estimator. The output of cross_val_score is a list.
Because RMSE is not directly available, MSE is used.
cross_val_score(estimator, X, y, cv=10, scoring='neg_mean_squared_error')
This gives the list of the opposite sign of the MSE, therefore we need to multiply -1 before taking the square root. The discussion about this issue.
GridSearchCV
GridSearchCV fits the models with different parameters. A dictionary of names of parameters and values is called a parameter grid. The following is an example of the case of KNN.
param_grid = {
'n_neighbors' : range(1,31),
'weights' : ['uniform', 'distance'],
} # parameter grid
After that we create an instance of an estimator (without forgetting to import the module for the prediction), we instantiate the grid.
from sklearn.model_selection import GridSearchCV
grid = GridSearchCV(KNeighborsClassifier(), param_grid, cv=5, scoring='accuracy')
We can specify the number of CPUs for the computation by the option n_jobs. (-1 means all.)
grid.fit(X, y) # fit the grid with data
grid.grid_scores_: the complete results of validation scoresgrid.best_params_: the estimated parameter with the best CV scoregrid.best_score_: the best CV scoregrid.best_estimator_: the trained model with the best parameter- We can directly use the best model by
grid.predict()for prediction.
RandomizedSearchCV
RandomizedSearchCV is very similar to GridSearchCV, but the (combinations of) parameters are randomly chosen. A "parameter distributions" is used rather than a "parameter grid":
param_dist = {
'n_neighbors' : range(1,31),
'weights' : ['uniform', 'distance']
}
The difference is: we can specify a continuous distribution (rather than a list of values) for a continous parameters. (See the document for more details.)
from sklearn.grid_search import RandomizedSearchCV
rand = RandomizedSearchCV(knn, param_dist, cv=10, scoring='accuracy', n_iter=10, random_state=5)
Here knn is (an example of) an instance of an estimator. n_iter is the number of (combinations of) sampled parameters. random_state is the seed of random values.
After fitting rand.fit(X,y), the following attributes give the results.
rand.grid_scores_ # the all results (list of named tuples)
rand.best_params_ # the parameters giving the best accuracy (dict)
rand.best_score_ # the best accuracy
We may use the estimator of the best model through rand.best_estimator_ (the instance of the estimator).
Create an original estimator
from sklearn.base import BaseEstimator, ClassifierMixin
from collections import Counter
class MajorityVote(BaseEstimator, ClassifierMixin):
def __init__ (self,answer=0):
self.answer = answer
def fit(self,X,y):
val_count = Counter(y)
vals = sorted(val_count,key=lambda x: val_count[x], reverse=True)
self.answer = vals[0]
return self
def predict(self,X):
y = np.ones((X.shape[0]))
y *= self.answer
return y
